Genetic analysis of microbiomes from two 4,000-year-old teeth shows notable changes in the oral microenvironment since the Bronze Age, especially following the widespread consumption of sugar, according to research published in Molecular Biology and Evolution.
Despite the teeth being from the same person, they showed distinct genomes for Tannerella forsythia (T. forsythia), a bacteria associated with gum disease. Furthermore, other ancient genomes have shed light on human history. Streptococcus mutans (S. mutans), a major contributor to tooth decay, has yet to be identified in preserved samples. However, the teeth, discovered in an Irish limestone cave, contained S. mutans, suggesting significant shifts in oral microbiota throughout history, the authors wrote.
“These results highlight that recent cultural transitions, such as the popularization of sugar, are most relevant to understanding the shaping of present-day oral pathobiont diversity,” wrote the authors, led by Lara Cassidy, PhD, an assistant professor at the Trinity College Dublin School of Genetics and Microbiology in Ireland (Mol Biol Evol, March 3, 2024).
Following the discovery, the two teeth underwent ancient DNA analysis, and it was determined they both belonged to the same adult male who lived between 2,280 to 2,140 cal BC. Both teeth were sampled twice, from the root and the crown. Metagenomic profiles of these samples were assessed and compared with publicly available ancient sequence data from dental calculus and teeth, the authors wrote.
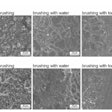
The team reconstructed ancient genomes for T. forsythia and S. mutans, revealing significant changes in oral bacteria populations and functions over the last 750 years. They found distinct differences in virulence factors between preindustrial and postindustrial genomes, which were especially pronounced in T. forsythia.
These changes likely affected the host immune response and interactions with other oral microbes. While only one ancient S. mutans genome was analyzed, it suggests that the modern mutacin repertoire is a recent acquisition, they wrote.
Both S. mutans and T. forsythia populations expanded after the medieval period, with S. mutans showing deeper diversity. T. forsythia experienced a rapid population increase about 600 years ago, coinciding with the rise of sugar in diets. S. mutans, known for thriving in high-sucrose environments, utilizes sucrose to create glucans, aiding in biofilm formation and maintaining a low pH, which gives it an acid-tolerant advantage, according to the study.
In the future, enhanced sampling of ancient microbial populations could yield fresh insights into the evolutionary processes governing bacterial taxon development, adaptation, and diversity maintenance, contingent upon species and environmental factors, Cassidy et al added.
“Overall, our findings demonstrate the utility of ancient genomes in characterizing different modes of pathobiont evolution,” they concluded.