European researchers have identified unique DNA markings that may predict the risk of developing head and neck cancer, according to a study in the journal Cancer. The discovery may lead to the development of noninvasive tests for early precancer screening, allowing at-risk patients to get earlier treatment and improve their survival.
Nearly 42,000 Americans will be diagnosed with oral or pharyngeal cancer this year, resulting in more than 8,000 deaths, according to the Oral Cancer Foundation. Of the newly diagnosed patients, about 57% will be alive in five years. Patients with advanced oral cancer have poor survival rates (10% to 30% at five years).
Survival rates have not significantly improved in decades; oral cancer fatality rates are higher than cervical cancer and Hodgkin's lymphoma.
In the U.K., about 16,000 people are diagnosed annually with head and neck cancer, according to researchers from Queen Mary University of London. Worldwide, more than 640,000 new oral cancer cases are diagnosed annually, and global figures for oral cancer are estimated to rise to more than 1 million a year by 2030, according to the World Health Organization.
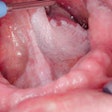
Oral lesions are very common, but only 5% to 30% may develop into carcinomas. If the carcinomas are detected in the early stages, treatment can be curative, but no test has been able to accurately detect which lesions will become cancerous.
 Muy-Teck Teh, PhD.
Muy-Teck Teh, PhD.
Currently, the diagnostic gold standard is histopathology, a relatively invasive procedure. Many oral cancers are now diagnosed at later stages when the chances of survival are significantly reduced.
A test previously developed by a team of British dental researchers led by Muy-Teck Teh, PhD, of the Institute of Dentistry at Queen Mary University of London showed cancer-detection accuracy rates of 91% to 94% when used on more than 350 head and neck tissue specimens from 299 patients in the U.K. and Norway. They published a study explaining the test in 2012 in International Journal of Cancer.
Their research on the cancer gene FOXM1 showed that when it is overexpressed the protein loses its control over cell growth, allowing cells to proliferate abnormally.
The researchers had identified a panel of 30 FOXM1-driven differentially methylated genes, a subset of which was found to be differentially expressed in head and neck squamous cell carcinoma (HNSCC) cells. Of the 30 candidate genes, they identified eight putative genes (hypomethylated: SPCS1, FLNA, CHPF, and GLT8D1; hypermethylated: C6orf136, MGAT1, NDUFA10, and PAFAH1B3) showing differential gene expression in a small patient cohort.
In the current study (Cancer, October 1, 2013), Teh and colleagues from the U.K. and Norway sought to further validate the eight genes by measuring their expression in a larger clinical sample set involving two geographically independent patient cohorts.
Methodology
The study tested two patient groups from Norway and the U.K. The Norwegian group consisted of retrospective frozen archival biopsies donated by 126 patients (histopathologic classification: 26 unaffected, 35 oral lichen planus [OLP], and 65 head and neck squamous cell carcinoma [HNSCC] patients). The unaffected normal oral tissue specimens were donated by healthy patients undergoing third-molar extractions. For the U.K., group, 28 patients prospectively provided paired margin (taken from unaffected tissue adjacent to the tumor from the same patient) and HNSCC tumor core tissue biopsies.
Sex and age distributions in the two groups were similar: 58% were male in the Norway cohort; 56% were male in the U.K. group; median age was 56 years for Norway and 59 years for the U.K. All of the Norwegians were white; in the U.K. group, 61% were white and 36% were Asian. Some 34% of the Norwegians and 32% of the U.K. group used tobacco.
In the Norwegian group, patients' tumors originated mainly from the tongue (25%), gingiva (23%), and larynx (23%). The U.K. cancer patients' tumors originated mainly from buccal (32%) and tongue (25%) tissue.
The researchers used their molecular diagnostic test, the Quantitative Malignancy Index Diagnostic System (qMIDS) assay, to further validate the specimens and verify their molecular malignancy status. The test measures the levels of 16 genes, which are converted via a diagnostic algorithm into a "malignancy index" that quantifies the risk of the lesion becoming cancerous. It is less invasive than the standard histopathology methods because it requires only a 1- to 2-mm piece of tissue, and it takes less than three hours for results, compared with up to a week for standard histopathology, according to the researchers.
In the Norwegian group, normal samples produced a median qMIDS malignancy index score of 1.0 (mean ± standard deviation [SD], 1.0 ± 0.1); OLP samples were scored 2.2 (2.2 ± 0.9), and HNSCCs were 9.1 (9.3 ± 1.7). In the U.K. group, unaffected margin samples had a median score of 1.3 (2.0 ± 1.9) and HNSCCs had a score of 8.9 (8.9 ± 2.2).
Gene expression analysis of the eight genes showed heterogeneous mRNA expression in both patient cohorts compared with controls, according to the researchers. Despite the heterogeneity, three of the four candidate hypomethylated genes tended to show upregulation in both HNSCC patient cohorts.
"DNA methylation is one of the fundamental epigenetic programming mechanisms whereby its heritable yet highly dynamic methylome landscape is able to regulate diverse phenotypes without altering the primary sequence of genomic DNA," the researchers wrote. Aberrant disturbance of the methylome landscape in normal cells is known to induce cancer.
Conclusion
The researchers identified and verified that FLNA, CHPF, and GLT8D1 were upregulated in HNSCC, and C6orf136 was downregulated in HNSCC in the two independent patient cohorts. Their differential gene expression was inversely correlated with differential promoter methylation, indicating that these genes were epigenetically modified in HNSCC tumors.
"Identification of these epigenetically regulated oncogenes and tumor-suppressor genes may be exploitable for clinical use as early biomarkers of cancer predisposition," they concluded. "An advantage of epigenetic DNA markers is that it enables the possible use of noninvasive specimens, such as saliva, buccal scrapes, and blood serum, for early cancer screening, diagnosis, and prognosis."
However, further study is needed, as the researchers are at the "discovery stage at the moment and have not used this as a diagnostic test as yet," they noted. But, "the eventual aim would be to test asymptomatic patients and/or people with unknown mouth lesions."