An interactive teaching tool being developed at the Creighton University School of Dentistry could improve nonradiologists' interpretation of cone-beam CT (CBCT) images, according to a presentation last week at the American Academy of Oral and Maxillofacial Radiology (AAOMR) annual conference in San Diego.
"There are 3,000 to 4,000 cone-beam CT systems in the U.S., with many of them in dental offices," noted Douglas Benn, DDS, PhD, a professor and director of oral and maxillofacial radiology at Creighton. "However, most of the users have not been trained to read CT anatomy."
Given that only 120 or so oral and maxillofacial radiologists are in the U.S., "novel methods are needed to expand radiographic teaching," he added.
“Until you can recognize normal, you cannot recognize abnormal.”
— Douglas Benn, DDS, PhD, Creighton
University
While some interactive teaching programs are available, they are usually limited to only a single set of images. To provide a range of anatomy and navigational skills for image stacks such as those in a single CBCT scan, a flexible program that can input multiple patient CT scans is needed, according to Dr. Benn.
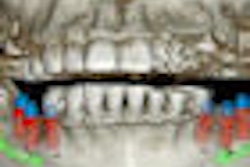
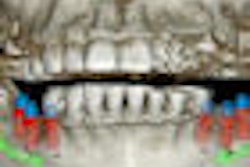
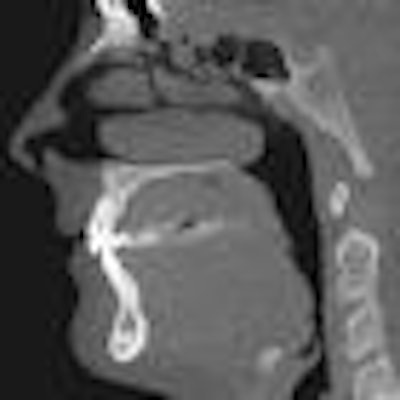
This challenge prompted him to develop a software tool that automates the reading of dental CT scans by recognizing and labeling key anatomical features. Using the National Institutes of Health's ImageJ Java programming tool and 1,000 lines of Java code, he wrote a platform-independent program that displays a window containing a stack of 512 axial head CT slices, a menu of 100 different anatomical sites, an example of a labeled slice for each anatomical site, and a diagram of the anatomy.
The user is asked to navigate through the stack of images to find the required anatomical object and click on it. The site is automatically labeled, and the x,y,z coordinates are stored for later examination to confirm that the selected anatomy is correct.
The program can load a stack of 512 images in about 15 seconds, Dr. Benn noted. With minimal training, it takes a user about 30 minutes to label 100 sites.
The challenge now, he said, is to load in some 300 patient cases to create a library of patient datasets. Once all the anatomy in all of the slices is labeled, it will be possible to automatically grade student selections, Dr. Been said.
"You need an enormous amount of data to get a range of normal," he said. "Until you can recognize normal, you cannot recognize abnormal."
The development effort is part of a larger artificial intelligence program at Creighton designed to automate CT training, noted Dr. Benn, who has been doing computer programming since 1980 and has a master's degree in computer science and artificial intelligence.
"Why are we doing all this?" he said. "Because there is a shortage of experts in the world, and it is very expensive and time-consuming to train people. This software will reduce the amount of time it takes to train radiologists to read images."
Copyright © 2010 DrBicuspid.com